Abstract
Introduction
Patients with Sickle cell disease (SCD) have lower risk for HIV-1 infection. We showed that ex vivo HIV-1 replication is blocked in SCD PBMCs in part because of the increased expression of ferroportin (FPN) and activation of SAMHD1, a host antiviral restriction factor. We hypothesized that rupture of sickling red blood cells releases sickle cell hemoglobin (HbS) that is phagocyted by macrophages leading to upregulation of innate antiviral response and inhibition of HIV-1 replication. We accessed changes in antiviral gene expression in PBMCs obtained from SCD patients compared to healthy controls. We also analyzed antiviral gene expression in macrophages treated with HbS compared to the HbA treatment.
Methods
The study was approved by Howard University review board (IRB) and all subjects consented to sample collection. Whole blood was collected from 9 SCA patients and 9 age and gender- matched healthy controls. PBMCs were activated with PHA (0.5 μg/ml) for 24-48 hrs followed by IL-2 (10 U/ml) for 24 hrs. Human THP-1 cells were differentiated into macrophages with PMA (25nM) for 72 hrs and treated with purified HbS or HbA (5µM). RNA strand-specific libraries were constructed using TruSeq Stranded Total RNA Gold kit (Illumina) and sequenced on an Illumina NextSeq 500 using 75 bp paired-end sequencing on two v2.5 150 cycle High-Output kits, generating 40-50 million paired-end reads per sample. The sequencing data were mapped using Dragen RNA and compared using Dragen differential expression software (Illumina). Ingenuity Pathway analysis (IPA, Qiagen) was used for pathway analysis.
Results
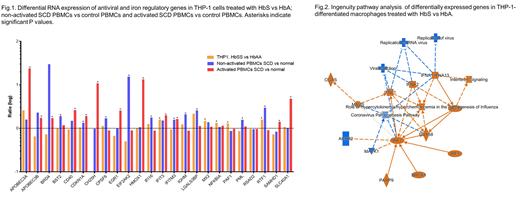
In activated SCD PBMCs compared to control PBMCs, 40432 genes were detected including 2230 differentially expressed genes (5.5%, 1.5-fold difference, 287 down and 1943 up) at 5% false discovery rate. In non-activated SCD PBMCs compared to control PBMCs, 33119 genes were detected including 5299 differentially expressed genes (16%, 923 down and 4376 up). In THP-1-differentiated macrophages treated with HbS versus HbA, 28362 genes were detected including 322 differentially expressed genes (1.1%, 187 down and 135 up).
We focused our analysis on 61 genes including viral restriction factors and iron regulatory genes. In activated SCD PBMCs, four genes had highest upregulation: APOBEC3A (23-fold, p=2 x 10 -5), CH25H (11-fold, p=4 x 10 -5), heme oxygenase-1 (HMOX1, 13-fold, p=1.5 x 10 -12) and FPN (SLC40A1, 5-fold, p = 9 x 10 -8) (Fig.1). Several additional genes were upregulated with 1.5-3-fold increase and high significance including APOBEC3B, BRD4, CD40, CDKN1A (p21), GDR1, IFIT3, IFITM3 and SAMHD1. In non-activated SCD PBMCs, the most upregulated gene was PKR (EIF2AK2, 15-fold, p=1 x 10 -11) and genes with 1.5-3 fold upregulation included APOBEC3B, BST2, CPSF6, IFI16, IFITM3, ISG15, LGALS3BP, PML and RTF1 (Fig.1). Of these genes, only IFITM3 overlapped between activated and non-activated PBMCs.
To test whether circulating HbS leads to the upregulation of antiviral response, we analyzed HbS-treated macrophages and found upregulation of several antiviral restriction factors (1.5-2.3 fold): IFIT3, LGALS3BP, MX2 and RTF1 (Fig.1). Unsupervised IPA showed upregulation of IRF-7 signaling pathway and down regulation of viral infection and replication (Fig.2). We validated the CH25 and HO-1 antiviral role in activated SCD PBMCs using small molecule inhibitors. We also confirmed overexpression of CH25H and HO-1 by western blot and ELISA. We observed higher levels of IRF7 in the activated SCD PBMCs confirming that it may play a role in the induction of antiviral response.
Conclusion
We propose that HbS released by hemolysis and uptaken by macrophages leads to the IRF-7-triggered induction of antiviral state in macrophages that will induced antiviral state in non-activated circulating PBMCs likely though the cytokines and interferons secretion known to be elevated in SCD patients. Upregulation of PKR (EIFAK2) levels in non-activated PBMCs strongly argue toward this possibility. Upon activation of PBMCs, additional factors are expressed including CH25H, HO-1, APOBEC3A and FPN that facilitated stronger and more robust anti-HIV-1 effect and block viral replication. Taken together, our study point to novel mechanism of upregulation of antiviral factors mediated by sickle cell hemoglobin that included induction of antiviral, heme- and iron- regulatory pathways.
No relevant conflicts of interest to declare.